Motivation
More than 600 disorders afflicting the nervous system is currently known. Neurodegenerative diseases are defined as hereditary and sporadic conditions which are characterized by progressive nervous system dysfunction. Current imaging device have abilities to see the tissue. Unfortunately evaluation of these data is time-consuming and often inaccurate.
Challenge Definition
This challenge deals with automatic measurement of brain volume taken from imaging devices with different settings, which can help with better diagnosis and more accurate treatment evaluation in future.
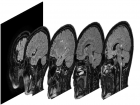
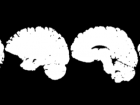
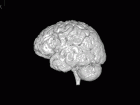
The objective of this challenge is to propose an automatic algorithm, which will transform the original set of images (see the figure below) into such an images, where the brain area is marked with white color and the rest is marked with black color. The results are compared to data labelled by human expert and can be evaluated by script that can be downloaded below.
Training and Testing Data
Although there are many methods for Goal of this challenge is selection brain tissue from MRI images. Each images set of a patient consists of 257 slices with resolution of 400 x 400 px and 16 bit grayscale bit depth.
- Download training and testing data.
- Download code for performance evaluation (JAVA): source code, binary.
| Experiment | ACCURACY | Description |
|---|---|---|
| Human expert | 99.540 % + 0.07756 | This express the accuracy that has been achieved by an independent human expert. |
| Threshold | 95.827 % +/- 0.8043 | This is naive method, where to each image slice was applied a simple threshold. |
Task illustration.
Source files
Training set contains scans of 12 patients with labels.
Testing set contains scans of 10 patients without labels.
Example solution created by thresholding.
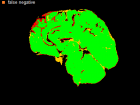
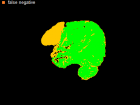
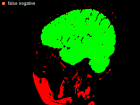
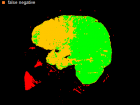
Errors of Human Expert and Threshold method
Examples of Human expert and threshold segmentation results.
How to evaluate
You can use our evaluator to check performance. This program simply calculates pixel error.
How to use from command line:
java -jar evaluator.jar D:\store\400_400\reference D:\store\400_400\humanExpert
Folders reference and humanExpert contains folders with patients’ masks. Or you just double click and GUI will appear.
Why to participate
- You will participate in still up-to-date research problem,
- You will be invited* to present your work on recognized TSP conference, which is indexed by most indexes, such as: IEEE Xplore®, CPCI (ISI web of Knowledge), SCOPUS, DBLP, and Google Scholar. More information about deadlines and information for authors can be found on page website: http://tsp.vutbr.cz/.
- Selected results will be sent to selected journal with Impact Factor.
* It is possible that some invited papers may be rejected. However, this is expected to be a rare case since the Organizer should recruit only the high-quality participants to the Special Session.
Citation
The training and testing data were first published in the paper:
UHER, V.; BURGET, R. Automatic 3D Segmentation of Human Brain Images Using Data- mining Techniques. In Proceedings of the 35th International Conference on Telecommunications and Signal Processing – TSP’ 2012. 1. 2012. p. 578-580. ISBN: 978-1-4673-1116- 8.
Contact
Ing. Radim Burget, Ph.D., burgetrm@feec.vutbr.cz